End-to-End Data Analysis Project: Part 2. Parsing the Headers
Sonny Torres, Thu 18 April 2019, Python
Sonny Torres, Thu 18 April 2019, Python
Contents
In my previous post I explained and showed how I scraped CMBS loan data from an HTML file downloaded from the SEC.gov website in order to refine my web scraping skills and acquire a dataset that was copyright free. A practical advantage of this dataset is that it is incredibly messy and often data "in the wild" requires a number of formatting steps before any analysis can be done.
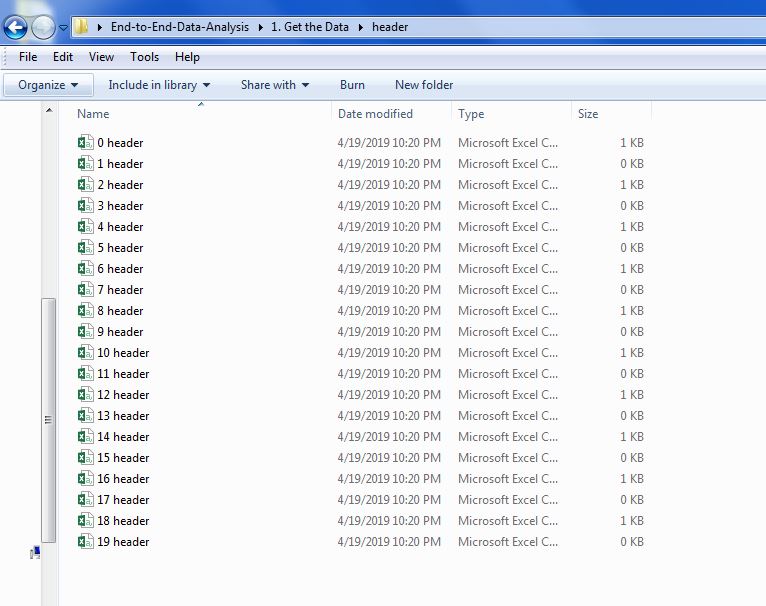
After the numerous HTML tables were scraped for their header data and table data, I saved each table and table header to a CSV file. Now I have two folders, one for headers and one for tables, full of csv files. Below is an image of the "headers" folder the script in my previous post created as well as all of the header files that were moved to it.
Below is all of the code that parses the header data and that I will be covering in this blog post.
from collections import Counter
import unicodedata
import numpy as np
import os
import csv
import pandas as pd
import glob
import re
main_dir = r"C:\Users\Username\Desktop\End to End Data Analysis Project\header"
os.chdir(main_dir)
pattern = '*header.csv'
all_header_files = [os.path.basename(file) for file in glob.glob(os.path.join(main_dir, pattern))]
def is_word(value):
if re.search(pattern=r'\w', string=value):
return True
else:
return False
def load_headers(filename):
f = open(filename)
reader = csv.reader(f)
header_rows = 0
header_dict = {}
for line in reader:
new_line = [unicodedata.normalize('NFKD', value) for value in line]
header_dict[header_rows] = new_line
header_rows += 1
main_header = header_dict[len(header_dict)-1]
main_header_len = len(main_header)
for row_ind, row in header_dict.items():
counts = Counter(row)
for word in enumerate(row):
word_index = word[0]
word_text = word[1]
if is_word(word_text) and counts[word_text] > 1:
header_dict[row_ind][word_index] = word_text + '_' + str(np.random.randint(10))
else:
continue
filename = filename.strip('.csv') + '_' + 'parsed' + '.csv'
with open(filename, 'w', newline='') as f:
writer = csv.writer(f)
writer.writerows(header_dict.values())
df = pd.read_csv(filename, encoding='ISO-8859-1', header=[number for number in range(header_rows)],
na_values='empty')
df.columns = [' '.join(col).strip() for col in df.columns.values]
return df
all_header_values = []
for num in range(0, 20, 2):
file = str(num) + ' header.csv'
df = load_headers(file)
all_header_values.append(df)
frame = pd.concat([value for value in all_header_values], axis=1)
print(frame)
frame.to_csv('frame.csv', index=False)
Below are the python libraries necessary to run the script. The main_dir variable is the path to the folder that contains the CSV header files as shown in the image above. I then used the os.chdir method from the os module to change to that directory. This is just a convenient way that I like to direct my python scripts to the correct folder but filepaths can be handled in many different ways.
from collections import Counter
import unicodedata
import numpy as np
import os
import csv
import pandas as pd
import glob
import re
main_dir = r"C:\Users\Username\Desktop\End to End Data Analysis Project\header"
os.chdir(main_dir)
pattern = '*header.csv'
all_header_files = [os.path.basename(file) for file in glob.glob(os.path.join(main_dir, pattern))]
def is_word(value):
if re.search(pattern=r'\w', string=value):
return True
else:
return False
Because I am only interested in files that end with the word "header" and have the file extension ".csv", I use *header.csv, the asterisk being a character used in the Regular Expressions library which means that the filename can start with anything as long as it ends with "header.csv".
The pattern and all_header_files variables are not necessary in this case because the folder only contains header files and os.chdir changed the current directory to the folder with the files but I included these variables and their explanations below because this is a common file manipulation technique that I have found very useful.
The header files will be contained in the list all_header_files. The os.path.join method simply joins the path provided, main_dir, and the pattern header.csv. Basically the join method appends "header.csv" to the main folder path. In order to utilize the pattern the way I wanted, that is, to catch any file that ends with "header.csv", I used the glob library to expand this pattern to generalize to all files that match the pattern. The list comprehension creates a list of each file's absolute path and os.path.basename extracts only the filename and extension from the absolute path. Again, a simple for loop to iterate over each file would suffice in this case, but I included the pattern and all_header_files as a reference for myself and others when looking to do common file manipulation operations. See the os.path documentation for more.
is_word, as you might have guessed, searches for words in a given string. The raw character string r'\w' uses the Regular Expressions module to search for any alphanumeric character.
The remainder of this blog post will cover the bulk of the script, which is the load_headers function. This function loads in each CSV file, one at a time, parses and cleans the header data and saves the modified headers as a new file. This is necessary because the headers, when concatenated with the table data, would be horribly misaligned and would cascade down one to three rows if they were not parsed prior to joining the data.
The function load_headers first accepts a filename (for example "2 header.csv"). Below, the open function creates a file object to be read by the csv module with the .reader method.
def load_headers(filename):
f = open(filename)
reader = csv.reader(f)
Similar to my first post, I iterate over each line of the data one at a time, horizonatally, across each row of data and again utilize a dictionary data container to easily have access to any portion of the data (e.g. a certain row or a certain word in a given row). Each header row will have an index value associated with it and each header will be stored as a list in the header_dict dictionary.
header_rows = 0
header_dict = {}
Before iterating over each header row of each CSV file to create dictionaries of the header rows, I had to make changes to the text data's encoding.
The concept of encoding has always been elusive to me until I found this great article, written by the Stack Overflow CEO, Joel Spolsky.
Here are my oversimplified definitions of what I have learned about encoding.
Python's default encoding is UTF-8 but the SEC website is encoded with Windows-1252. This mismatch of encodings created frustrating problems I did not understand. I would parse the header data, export to CSV and find strange characters all over the CSV file when I opened it in excel. The root of the problem is succinctly illustrated below in a chart, specifically the highlighted portion, I found on this website.
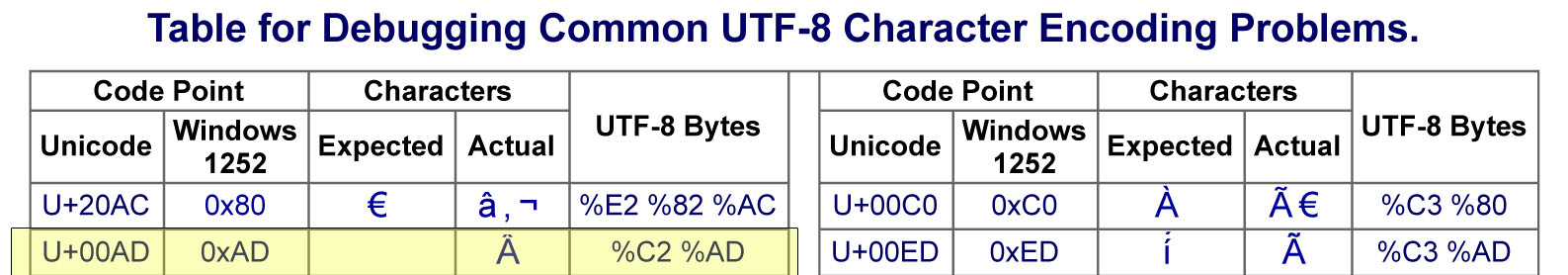
In the image above you can see there are not only different "inputs" (code points) but different outputs. The solution for this problem involved using the unicodedata library and its .normalize method. Using the chart's terminology, the 'NFKD' supplied value in the code below simply converts the actual character result with the expected character result under the Unicode standard. In this case, a blank space also known as a non-breaking space is the expected and desired value. This solution made it so that instead of a bunch of "ÂÂÂÂÂÂÂÂ"'s appearing in my excel file, they were blank cells like they were supposed to be before the encoding problem. The blank cells are important because they play an integral part in keeping the entire document's format consistent.
for line in reader:
new_line = [unicodedata.normalize('NFKD', value) for value in line]
header_dict[header_rows] = new_line
header_rows += 1
main_header = header_dict[len(header_dict)-1]
main_header_len = len(main_header)
The code above then creates the variable main_header, which is the bottom header row and its associated length. This bottom header row is important because some headers have text that spans down 2-3 rows while others do not and the bottom header row is the only consistent row that will be used to align other header rows above it.
The next section of the load_headers function involves iterating over the dictionary of header values, counting the instances of each word in a given header row and appending a random number from 0-9 to the word if it is not a unique word. This is important for a reason I will explain in the next couple of steps.
for row_ind, row in header_dict.items():
counts = Counter(row)
for word in enumerate(row):
word_index = word[0]
word_text = word[1]
if is_word(word_text) and counts[word_text] > 1:
header_dict[row_ind][word_index] = word_text + '_' + str(np.random.randint(10))
else:
continue
Below, ''filename'' is a variable that will store the new filename for the new CSV file that will contain the parsed header rows. filename is created by simply using the existing filename, stripping '.csv' from the end of the file, appending '_parsed' to the end and adding the '.csv' file extension back on the filename string.
filename = filename.strip('.csv') + '_' + 'parsed' + '.csv'
After the headers are normalized to Unicode encoding and the duplicates are accounted for, I use the common pythonic way of writing files, utilizing the with open technique to write the header_dict values (lists of header data) to a CSV file.
with open(filename, 'w', newline='') as f:
writer = csv.writer(f)
writer.writerows(header_dict.values())
Finally, I load the new CSV header file into a Pandas Dataframe for further editing. The reason I had to account for the duplicate values in the headers involves loading a MultiIndex with Pandas' ``read_csv`` method. Because I want to merge all of the header rows into a single header row, I need to be able to access the header rows and their values individually by loading the headers as a MultiIndex. The normal output of a MultiIndex is an array of tuples, which contain the string values for each row in a column. Below is an example of of how the Multindex is organized.
array([('col1_row1', 'col1_row2', 'col1_row3'),
('col2_row1', 'col2_row2', 'col2_row3'),
('col3_row1', 'col3_row2', 'col3_row3')])
This feature of Pandas is useful because you can access any given value you want by accessing the string values within the tuples. Utilizing the tuples for the MultiIndex was my intention with the code below.
df = pd.read_csv(filename, encoding='ISO-8859-1', header=[number for number in range(header_rows)],
na_values='empty')
My method of gluing together multiple header rows into one header row was achieved by iterating over each tuple value, putting a space in between each word and then stripping away the white space on the ends of each header by using the .strip method. Lastly, the load_headers function returns the dataframe with the cumulative changes made thus far.
df.columns = [' '.join(col).strip() for col in df.columns.values]
return df
However, when gluing the header rows together I noticed that my results for duplicate values came out strange. But only for duplicate values in a given row. My IPython session below illustrates why.
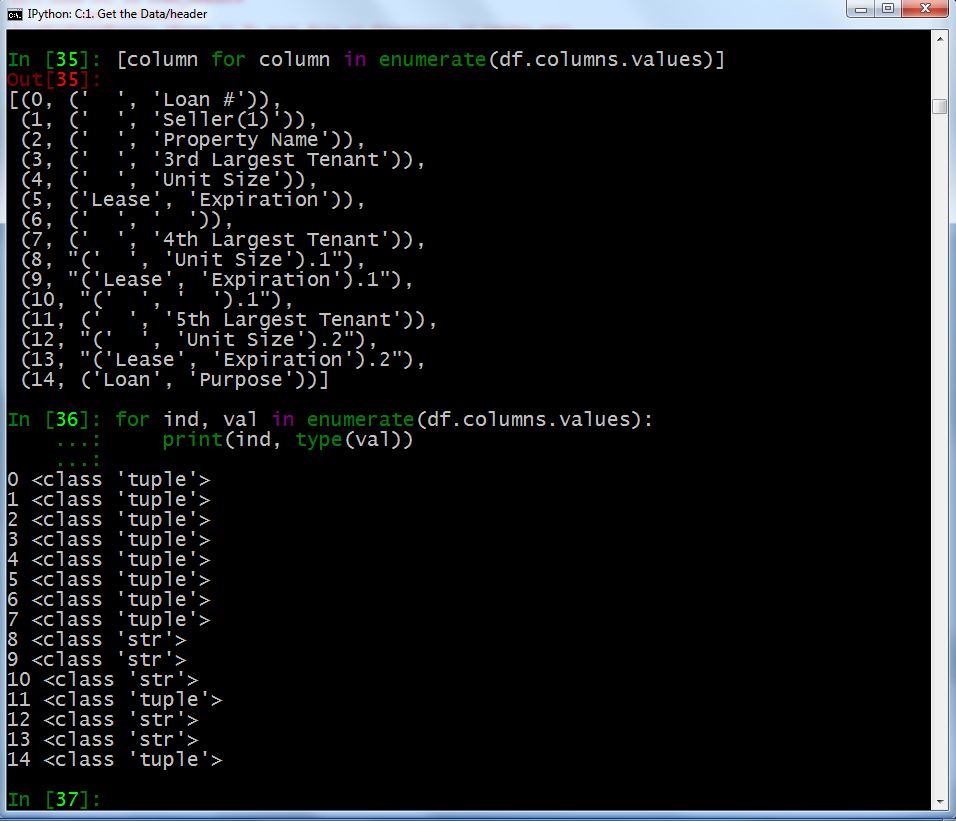
In the image above, you can see that the majority of header columns are tuples, as expected, but columns 8-10, 12 and 13 are string values. For reasons I do not know, Pandas will load duplicate values in a MultiIndex not as tuples but as string values. So, when I iterate over each header column like they are tuples as I did with the ' '.join(col)... list comprehension above, I ended up iterating over each character in those string values causing columns with duplicate values to look like this:
( ' L e a s e ' , ' E x p i r a t i o n ' ) . 1
In the output above, there is a space in between each letter caused by the ' '.join list comprehension. The iteration method worked fine on the column values that were tuples but created the undesired result when the column was actually a string value. Luckily, I found a way to differentiate the duplicate values by appending a random number as show earlier in this post.
Finally, a simple for loop iterates over the files in the directory, utilizes the load_headers function, appends each header file's headers to a single list all_header_values, concatenates the resulting list using the pd.concat method and saves the parsed headers into a single CSV file which I called 'frame.csv'
all_header_values = []
for num in range(0, 20, 2):
file = str(num) + ' header.csv'
df = load_headers(file)
all_header_values.append(df)
frame = pd.concat([value for value in all_header_values], axis=1)
print(frame)
frame.to_csv('frame.csv', index=False)
The reason I iterated over the range of numbers I did was because there were 19 files in the directory and I wanted to iterate over every other file because duplicate sets of headers were "skipped" by saving them as a blank CSV file as shown in the code in my first post.
Now that all the data is gathered and the headers have been parsed and combined into a single, neat row, the only thing left to produce a single table of data is to concatenate the table files and their respective headers, which I will do in my next post.